
 Star products
Star products
 Follow Us
Follow Us

Scan and follow us
Learn more
Tool | Function introduction | Provide products |
|---|---|---|
| KFERQ-PAmCherry1 | Detection of CMA activity | LV/Ad/AAV |
| PAmCherry1-KFERQ-NE |
KFERQ-PAmCherry1 and PAmCherry1-KFERQ-NE virus tools:
PAmCherry1 is a light-activated protein, which is lightless and needs to be activated by exposure to light at 405 nm for 5-10 minutes before it can be activated to emit red fluorescence (excitation 564 nm, emission 595 nm). The use of such fluorescent proteins reduces the effect of highly autofluorescent or cumulative autofluorescent pigment deposits on cellular observation. KFERQ-PAmCherry1 fuses to the photoactivated protein PAmCherry1 with the aid of the molecular chaperone recognition substrate motif KFERQ, which is converted into a CMA substrate, and when targeted to the lysosome, CMA activity can be detected in living cells by observing the number of red fluorescent spots.
Note: "NE" is a protein tag that can be used for protein expression detection.
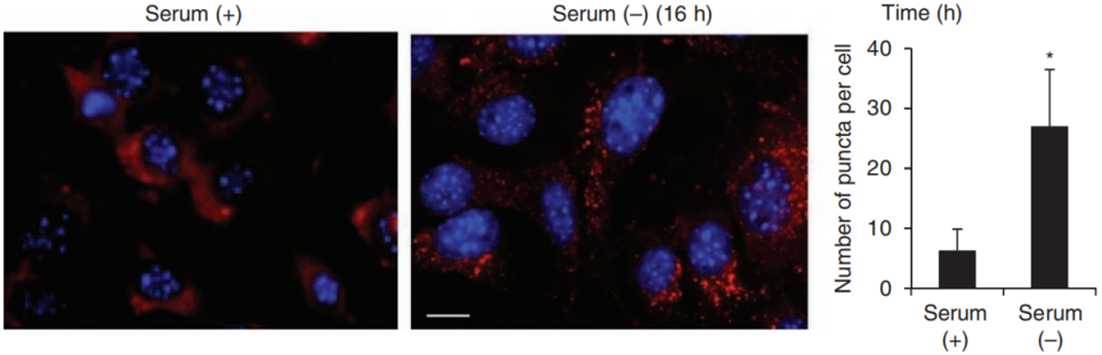
PMID: 21750540(KFERQ-PAmCherry1 tool)

PMID: 30983487(PAmCherry1-KFERQ-NE tool)

联系我们

返回顶部